Nitrogenase
Nitrogenase
Jump to navigation
Jump to search
| Nitrogenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC number | 1.18.6.1 | ||||||||
| CAS number | 9013-04-1 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Nitrogenases are enzymes (EC 1.18.6.1EC 1.19.6.1) that are produced by certain bacteria, such as cyanobacteria (blue-green algae). These enzymes are responsible for the reduction of nitrogen (N2) to ammonia (NH3). Nitrogenases are the only family of enzymes known to catalyze this reaction, which is a key step in the process of nitrogen fixation. Nitrogen fixation is required for all forms of life, with nitrogen being essential for the biosynthesis of molecules (nucleotides, amino acids) that create plants, animals and other organisms.
Contents
1 Structure
1.1 Fe protein
1.2 MoFe protein
2 Mechanism
2.1 General mechanism
2.2 Lowe-Thorneley kinetic model
2.3 Intermediates E0 through E4
2.4 Distal and alternating pathways for N2 fixation
2.5 Mechanism of MgATP binding
2.6 Other mechanistic details
3 Nonspecific reactions
4 Organisms that synthesize nitrogenase
5 Similarity to other proteins
6 Measurement of nitrogenase activity
7 See also
8 References
9 Further reading
10 External links
Structure[edit]
The equilibrium formation of ammonia from molecular hydrogen and nitrogen has an overall negative enthalpy of reaction (ΔH0=−45.2 kJmol−1NH3displaystyle Delta H^0=-45.2 mathrm kJ ,mathrm mol^-1 ;mathrm NH_3 

The nitrogenase complex consists of two proteins:
- The homodimeric Fe protein, a reductase which has a high reducing power and is responsible for the supply of electrons.
- The heterotetrameric MoFe protein, a nitrogenase which uses the electrons provided to reduce N2 to NH3.

Structure of the FeMo cofactor showing the sites of binding to nitrogenase (the amino acids cys and his).
Fe protein[edit]
The Fe protein is a dimer of identical subunits which contains one [Fe4S4] cluster and weighs approximately 60-64kDa.[2] The function of the Fe protein is to transfer electrons from a reducing agent, such as ferredoxin or flavodoxin to the MoFe protein. The transfer of electrons requires an input of chemical energy which comes from the binding and hydrolysis of ATP. The hydrolysis of ATP also causes a conformational change within the nitrogenase complex, bringing the Fe protein and MoFe protein closer together for easier electron transfer.[citation needed]
MoFe protein[edit]
The MoFe protein is a heterotetramer consisting of two α subunits and two β subunits, weighing approximately 240-250kDa.[2] The MoFe protein also contains two iron-sulfur clusters, known as P-clusters, located at the interface between the α and β subunits and two FeMo cofactors, within the α subunits. The oxidation state of Mo in these nitrogenases was formerly thought Mo(V), but more recent evidence is for Mo(III).[3] (Molybdenum in other enzymes generally resides in the molybdenum cofactor, as fully oxidized Mo(VI)).
- The core (Fe8S7) of the P-cluster takes the form of two [Fe4S3] cubes linked by a central sulfur atom. Each P-cluster is covalently linked to the MoFe protein by six cysteine residues.
- Each FeMo cofactor (Fe7MoS9C) consists of two non-identical clusters: [Fe4S3] and [MoFe3S3], which are linked by three sulfide ions. Each FeMo cofactor is covalently linked to the α subunit of the protein by one cysteine residue and one histidine residue.
Electrons from the Fe protein enter the MoFe protein at the P-clusters, which then transfer the electrons to the FeMo cofactors. Each FeMo cofactor then acts as a site for nitrogen fixation, with N2 binding in the central cavity of the cofactor.
Mechanism[edit]

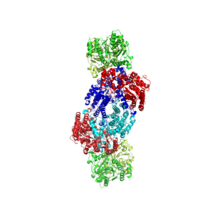
Figure 1: Nitrogenase with key catalytic sites highlighted. There are two sets of catalytic sites within each nitrogenase enzyme.

Figure 2: Nitrogenase with one set of metal clusters magnified. Electrons travel from the Fe-S cluster (yellow) to the P cluster (red), and end at the FeMo-co (orange).
General mechanism[edit]

Figure 3: Key catalytic sites within nitrogenase. Atoms are colored by element. Top: Fe-S Cluster Middle: P Cluster Bottom: FeMo-co
Nitrogenase is an enzyme responsible for catalyzing nitrogen fixation, which is the reduction of nitrogen (N2) to ammonia (NH3) and a process vital to sustaining life on Earth.[4] There are three types of nitrogenase found in various nitrogen-fixing bacteria: molybdenum (Mo) nitrogenase, vanadium (V) nitrogenase, and iron-only (Fe) nitrogenase.[5] Molybdenum nitrogenase, which can be found in diazotrophs and legumes,[6][7] is the nitrogenase that has been studied the most extensively and thus is the most well characterized.[5] Figures 1-2 display the crystal structure and key catalytic components of molybdenum nitrogenase extracted from Azotobacter vinelandii. Vanadium nitrogenase and iron-only nitrogenase can both be found in select species of Azotobacter as an alternative nitrogenase.[6][8] Equations 1 and 2 show the balanced reactions of nitrogen fixation in molybdenum nitrogenase and vanadium nitrogenase respectively.
N2 + 8 H+ + 8 e− + 16 MgATP → 2 NH3 + H2 + 16 MgADP + 16 Pi[4]
(1)
N2 + 14 H+ + 12 e− + 40 MgATP → 2 NH4+ + 3 H2 + 40 MgADP + 40 Pi[9]
(2)
All nitrogenases are two-component systems made up of Component I (also known as dinitrogenase) and Component II (also known as dinitrogenase reductase). Component I is a MoFe protein in molybdenum nitrogenase, a VFe protein in vanadium nitrogenase, and a Fe protein in iron-only nitrogenase.[4] Component II is a Fe protein that contains the Fe-S cluster (Figure 3: top), which transfers electrons to Component I.[8] Component I contains 2 key metal clusters: the P-cluster (Figure 3: middle), and the FeMo-cofactor (FeMo-co) (Figure 3: bottom). Mo is replaced by V or Fe in vanadium nitrogenase and iron-only nitrogenase respectively.[4] During catalysis, electrons flow from a pair of ATP molecules within Component II to the Fe-S cluster, to the P-cluster, and finally to the FeMo-co, where reduction of N2 to NH3 takes place.
Lowe-Thorneley kinetic model[edit]
The reduction of nitrogen to two molecules of ammonia is carried out at the FeMo-co of Component I after the sequential addition of proton and electron equivalents from Component II.[4]Steady state, freeze quench, and stopped-flow kinetics measurements carried out in the 70’s and 80’s by Lowe, Thorneley, and others provided a kinetic basis for this process.[10][11] The Lowe-Thorneley (LT) kinetic model (depicted in Figure 4) was developed from these experiments and documents the eight correlated proton and electron transfers required throughout the reaction.[4][10][11] Each intermediate stage is depicted as En where n = 0-8, corresponding to the number of equivalents transferred. The transfer of four equivalents are required before the productive addition of N2, although reaction of E3 with N2 is also possible.[10] Notably, nitrogen reduction has been shown to require 8 equivalents of protons and electrons as opposed to the 6 equivalents predicted by the balanced chemical reaction.[12]
Intermediates E0 through E4[edit]
Spectroscopic characterization of these intermediates has allowed for greater understanding of nitrogen reduction by nitrogenase, however, the mechanism remains an active area of research and debate. Briefly listed below are spectroscopic experiments for the intermediates before the addition of nitrogen:
E0 – This is the resting state of the enzyme before catalysis begins. EPR characterization shows that this species has a spin of 3/2.[13]
E1 – The one electron reduced intermediate has been trapped during turnover under N2. Mӧssbauer spectroscopy of the trapped intermediate indicates that the FeMo-co is integer spin greater than 1.[14]

Figure 4: Lowe-Thorneley kinetic model for reduction of nitrogen to ammonia by nitrogenase.
E2 – This intermediate is proposed to contain the metal cluster in its resting oxidation state with the two added electrons stored in a bridging hydride and the additional proton bonded to a sulfur atom. Isolation of this intermediate in mutated enzymes shows that the FeMo-co is high spin and has a spin of 3/2.[15]
E3 – This intermediate is proposed to be the singly reduced FeMo-co with one bridging hydride and one proton.[4]
E4 – Termed the Janus intermediate after the Roman god of transitions, this intermediate is positioned after exactly half of the electron proton transfers and can either decay back to E0 or proceed with nitrogen binding and finish the catalytic cycle. This intermediate is proposed to contain the FeMo-co in its resting oxidation state with two bridging hydrides and two sulfur bonded protons.[4] This intermediate was first observed using freeze quench techniques with a mutated protein in which residue 70, a valine amino acid, is replaced with isoleucine.[16] This modification prevents substrate access to the FeMo-co. EPR characterization of this isolated intermediate shows a new species with a spin of ½. ENDOR experiments have provided insight into the structure of this intermediate, revealing the presence of two bridging hydrides.[16]95Mo and 57Fe ENDOR show that the hydrides bridge between two iron centers.[17] Cryoannealing of the trapped intermediate at -20 °C results in the successive loss of two hydrogen equivalents upon relaxation, proving that the isolated intermediate is consistent with the E4 state.[4] The decay of E4 to E2 + H2 and finally to E0 and 2H2 has confirmed the EPR signal associated with the E2 intermediate.[4]
The above intermediates suggest that the metal cluster is cycled between its original oxidation state and a singly reduced state with additional electrons being stored in hydrides. It has alternatively been proposed that each step involves the formation of a hydride and that the metal cluster actually cycles between the original oxidation state and a singly oxidized state.[4]
Distal and alternating pathways for N2 fixation[edit]

Figure 5: Distal and alternating mechanistic pathways for nitrogen fixation in nitrogenase.
While the mechanism for nitrogen fixation prior to the Janus E4 complex is generally agreed upon, there are currently two hypotheses for the exact pathway in the second half of the mechanism: the "distal" and the "alternating" pathway (see Figure 5).[4][18][19] In the distal pathway, the terminal nitrogen is hydrogenated first, releases ammonia, then the nitrogen directly bound to the metal is hydrogenated. In the alternating pathway, one hydrogen is added to the terminal nitrogen, then one hydrogen is added to the nitrogen directly bound to the metal. This alternating pattern continues until ammonia is released.[4][18][19] Because each pathway favors a unique set of intermediates, attempts to determine which path is correct have generally focused on the isolation of said intermediates, such as the nitrido in the distal pathway,[20] and the diazene and hydrazine in the alternating pathway.[4] Attempts to isolate the intermediates in nitrogenase itself have so far been unsuccessful, but the use of model complexes has allowed for the isolation of intermediates that support both sides depending on the metal center used.[4] Studies with Mo generally point towards a distal pathway, while studies with Fe generally point towards an alternating pathway.[4][18][19][21][22]
Specific support for the distal pathway has mainly stemmed from the work of Schrock and Chatt, who successfully isolated the nitrido complex using Mo as the metal center in a model complex.[20][23] Specific support for the alternating pathway stems from a few studies. Iron only model clusters have been shown to catalytically reduce N2.[21][22] Small tungsten clusters have also been shown to follow an alternating pathway for nitrogen fixation.[24] The vanadium nitrogenase releases hydrazine, an intermediate specific to the alternating mechanism.[4][25] However, the lack of characterized intermediates in the native enzyme itself means that neither pathway has been definitively proven. Furthermore, computational studies have been found to support both sides, depending on whether the reaction site is assumed to be at Mo (distal) or at Fe (alternating) in the MoFe cofactor.[4][18][19]
Mechanism of MgATP binding[edit]

Binding of MgATP is one of the central events to occur in the mechanism employed by nitrogenase. Hydrolysis of the terminal phosphate group of MgATP provides the energy needed to transfer electrons from the Fe protein to the MoFe protein.[26] The binding interactions between the MgATP phosphate groups and the amino acid residues of the Fe protein are well understood by comparing to similar enzymes, while the interactions with the rest of the molecule are more elusive due to the lack of a Fe protein crystal structure with MgATP bound (as of 1996).[27] Three protein residues have been shown to have significant interactions with the phosphates, shown in Figure 6.[10] In the absence of MgATP, a salt bridge exists between residue 15, lysine, and residue 125, aspartic acid.[27] Upon binding, this salt bridge is interrupted. Site-specific mutagenesis has demonstrated that when the lysine is substituted for a glutamine, the protein’s affinity for MgATP is greatly reduced[28] and when the lysine is substituted for an arginine, MgATP cannot bind due to the salt bridge being too strong.[29] The necessity of specifically aspartic acid at site 125 has been shown through noting altered reactivity upon mutation of this residue to glutamic acid.[30] The third residue that has been shown to be key for MgATP binding is residue 16, serine. Site-specific mutagenesis was used to demonstrate this fact.[30] This has led to a model in which the serine remains coordinated to the Mg2+ ion after phosphate hydrolysis in order to facilitate its association with a different phosphate of the now ADP molecule.[31] MgATP binding also induces significant conformational changes within the Fe protein.[10] Site-directed mutagenesis was employed to create mutants in which MgATP binds but does not induce a conformational change.[32] Comparing X-ray scattering data in the mutants versus in the wild-type protein led to the conclusion that the entire protein contracts upon MgATP binding, with a decrease in radius of approximately 2.0 Å.[32]
Other mechanistic details[edit]
Many mechanistic aspects of catalysis remain unknown. No crystallographic analysis has been reported on substrate bound to nitrogenase. One problem is that the enzyme is generally obtained as a mixture of states.[citation needed] Nitrogenase is able to reduce acetylene, but is inhibited by carbon monoxide, which binds to the enzyme and thereby prevents binding of dinitrogen. Dinitrogen prevent acetylene binding, but acetylene does not inhibit binding of dinitrogen and requires only one electron for reduction to ethylene.[33] Due to the oxidative properties of oxygen, most nitrogenases are irreversibly inhibited by dioxygen, which degradatively oxidizes the Fe-S cofactors.[citation needed] This requires mechanisms for nitrogen fixers to protect nitrogenase from oxygen in vivo. Despite this problem, many use oxygen as a terminal electron acceptor for respiration.[citation needed] Although the ability of some nitrogen fixers such as Azotobacteraceae to employ an oxygen-labile nitrogenase under aerobic conditions has been attributed to a high metabolic rate, allowing oxygen reduction at the cell membrane, the effectiveness of such a mechanism has been questioned at oxygen concentrations above 70 µM (ambient concentration is 230 µM O2), as well as during additional nutrient limitations.[34]
Nonspecific reactions[edit]
In addition to dinitrogen reduction, nitrogenases also reduce protons to dihydrogen, meaning nitrogenase is also a dehydrogenase. A list of other reactions carried out by nitrogenases is shown below:[35][36]
HC≡CH → H2C=CH2
N–=N+=O → N2 + H2O
N=N=N– → N2 + NH3
C≡N−
→ CH4, NH3, H3C–CH3, H2C=CH2 (CH3NH2)- N≡C–R → RCH3 + NH3
- C≡N–R → CH4, H3C–CH3, H2C=CH2, C3H8, C3H6, RNH2
O=C=S → CO + H2S [37][38]
O=C=O → CO + H2O [37]
S=C=N– → H2S + HCN [38]
O=C=N– → H2O + HCN, CO + NH3[38]
Furthermore, dihydrogen functions as a competitive inhibitor,[39]carbon monoxide functions as a non-competitive inhibitor,[35][36] and carbon disulfide functions as a rapid-equilibrium inhibitor[37] of nitrogenase.
Vanadium nitrogenases have also been shown to catalyze the conversion of CO into alkanes through a reaction comparable to Fischer-Tropsch synthesis.
Organisms that synthesize nitrogenase[edit]
There are two types of bacteria that synthesize nitrogenase and are required for nitrogen fixation. These are:
- Free-living bacteria (non-symbiotic), examples include:
Cyanobacteria (blue-green algae)- Green sulfur bacteria
- Azotobacter
- Mutualistic bacteria (symbiotic), examples include:
Rhizobium, associated with leguminous plants
Spirillum, associated with cereal grasses- Frankia
Similarity to other proteins[edit]
The three subunits of nitrogenase exhibit significant sequence similarity to three subunits of the light-independent version of protochlorophyllide reductase that performs the conversion of protochlorophyllide to chlorophyll. This protein is present in gymnosperms, algae, and photosynthetic bacteria but has been lost by angiosperms during evolution.[40]
Separately, two of the nitrogenase subunits (NifD and NifH) have homologues in methanogens that do not fix nitrogen e.g. Methanocaldococcus jannaschii.[41] Little is understood about the function of these "class IV" nif genes,[42] though they occur in many methanogens. In M. jannaschii they are known to interact with each other and are constitutively expressed.[41]
Measurement of nitrogenase activity[edit]
As with many assays for enzyme activity, it is possible to estimate nitrogenase activity by measuring the rate of conversion of the substrate (N2) to the product (NH3). Since NH3 is involved in other reactions in the cell, it is often desirable to label the substrate with 15N to provide accounting or "mass balance" of the added substrate. A more common assay, the acetylene reduction assay or ARA, estimates the activity of nitrogenase by taking advantage of the ability of the enzyme to reduce acetylene gas to ethylene gas. These gases are easily quantified using gas chromatography.[43] Though first used in a laboratory setting to measure nitrogenase activity in extracts of Clostridium pasteurianum cells, ARA has been applied to a wide range of test systems, including field studies where other techniques are difficult to deploy. For example, ARA was used successfully to demonstrate that bacteria associated with rice roots undergo seasonal and diurnal rhythms in nitrogenase activity, which were apparently controlled by the plant.[44]
Unfortunately, the conversion of data from nitrogenase assays to actual moles of N2 reduced (particularly in the case of ARA), is not always straightforward and may either underestimate or overestimate the true rate for a variety of reasons. For example, H2 competes with N2 but not acetylene for nitrogenase (leading to overestimates of nitrogenase by ARA). Bottle or chamber-based assays may produce negative impacts on microbial systems as a result of containment or disruption of the microenvironment through handling, leading to underestimation of nitrogenase. Despite these weaknesses, such assays are very useful in assessing relative rates or temporal patterns in nitrogenase activity.
See also[edit]
- Nitrogen fixation
References[edit]
^ Modak JM (2002). "Haber Process for Ammonia Synthesis". Resonance. 7: 69–77. doi:10.1007/bf02836187..mw-parser-output cite.citationfont-style:inherit.mw-parser-output .citation qquotes:"""""""'""'".mw-parser-output .citation .cs1-lock-free abackground:url("//upload.wikimedia.org/wikipedia/commons/thumb/6/65/Lock-green.svg/9px-Lock-green.svg.png")no-repeat;background-position:right .1em center.mw-parser-output .citation .cs1-lock-limited a,.mw-parser-output .citation .cs1-lock-registration abackground:url("//upload.wikimedia.org/wikipedia/commons/thumb/d/d6/Lock-gray-alt-2.svg/9px-Lock-gray-alt-2.svg.png")no-repeat;background-position:right .1em center.mw-parser-output .citation .cs1-lock-subscription abackground:url("//upload.wikimedia.org/wikipedia/commons/thumb/a/aa/Lock-red-alt-2.svg/9px-Lock-red-alt-2.svg.png")no-repeat;background-position:right .1em center.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registrationcolor:#555.mw-parser-output .cs1-subscription span,.mw-parser-output .cs1-registration spanborder-bottom:1px dotted;cursor:help.mw-parser-output .cs1-ws-icon abackground:url("//upload.wikimedia.org/wikipedia/commons/thumb/4/4c/Wikisource-logo.svg/12px-Wikisource-logo.svg.png")no-repeat;background-position:right .1em center.mw-parser-output code.cs1-codecolor:inherit;background:inherit;border:inherit;padding:inherit.mw-parser-output .cs1-hidden-errordisplay:none;font-size:100%.mw-parser-output .cs1-visible-errorfont-size:100%.mw-parser-output .cs1-maintdisplay:none;color:#33aa33;margin-left:0.3em.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration,.mw-parser-output .cs1-formatfont-size:95%.mw-parser-output .cs1-kern-left,.mw-parser-output .cs1-kern-wl-leftpadding-left:0.2em.mw-parser-output .cs1-kern-right,.mw-parser-output .cs1-kern-wl-rightpadding-right:0.2em
^ ab Burges BK, Lowe DJ (1996). "Mechanism of Molybdenum Nitrogenase". Chemical Reviews. 96: 2983–3011. doi:10.1021/cr950055x. PMID 11848849.
^ Bjornsson R, Delgado-Jaime MU, Lima FA, Sippel D, Schlesier J, Weyhermüller T, Einsle O, Neese F, DeBeer S (2015). "Molybdenum L-Edge XAS Spectra of MoFe Nitrogenase". Z Anorg Allg Chem. 641: 65–71. doi:10.1002/zaac.201400446. PMC 4510703. PMID 26213424.
^ abcdefghijklmnopqr Hoffman BM, Lukoyanov D, Yang ZY, Dean DR, Seefeldt LC (April 2014). "Mechanism of nitrogen fixation by nitrogenase: the next stage". Chemical Reviews. 114 (8): 4041–62. doi:10.1021/cr400641x. PMC 4012840. PMID 24467365.
^ ab Peters JW, Szilagyi RK (April 2006). "Exploring new frontiers of nitrogenase structure and mechanism". Current Opinion in Chemical Biology. Bioinorganic chemistry / Biocatalysis and biotransformation. 10 (2): 101–8. doi:10.1016/j.cbpa.2006.02.019. PMID 16510305.
^ ab Rubio LM, Ludden PW (2008). "Biosynthesis of the iron-molybdenum cofactor of nitrogenase". Annual Review of Microbiology. 62 (1): 93–111. doi:10.1146/annurev.micro.62.081307.162737. PMC 3650356. PMID 18429691.
^ Franche C, Lindström K, Elmerich C (December 2008). "Nitrogen-fixing bacteria associated with leguminous and non-leguminous plants". Plant and Soil. 321 (1–2): 35–59. doi:10.1007/s11104-008-9833-8. ISSN 0032-079X.
^ ab Schneider K, Müller A (January 2004). Smith BE, Richards RL, Newton WE, eds. Catalysts for Nitrogen Fixation. Nitrogen Fixation: Origins, Applications, and Research Progress. Springer Netherlands. pp. 281–307. doi:10.1007/978-1-4020-3611-8_11. ISBN 978-90-481-6675-6.
^ Sippel D, Einsle O (2017-07-10). "The structure of vanadium nitrogenase reveals an unusual bridging ligand". Nature Chemical Biology. 13: 956–960. doi:10.1038/nchembio.2428.
^ abcde Burgess BK, Lowe DJ (November 1996). "Mechanism of Molybdenum Nitrogenase". Chemical Reviews. 96 (7): 2983–3012. doi:10.1021/cr950055x. PMID 11848849.
^ ab Wilson PE, Nyborg AC, Watt GD (July 2001). "Duplication and extension of the Thorneley and Lowe kinetic model for Klebsiella pneumoniae nitrogenase catalysis using a MATHEMATICA software platform". Biophysical Chemistry. 91 (3): 281–304. doi:10.1016/S0301-4622(01)00182-X. PMID 11551440.
^ Simpson FB, Burris RH (June 1984). "A nitrogen pressure of 50 atmospheres does not prevent evolution of hydrogen by nitrogenase". Science. 224 (4653): 1095–7. doi:10.1126/science.6585956. PMID 6585956.
^ Barney BM, Lee HI, Dos Santos PC, Hoffman BM, Dean DR, Seefeldt LC (May 2006). "Breaking the N2 triple bond: insights into the nitrogenase mechanism". Dalton Transactions (19): 2277–84. doi:10.1039/B517633F. PMID 16688314.
^ Yoo SJ, Angove HC, Papaefthymiou V, Burgess BK, Münck E (May 2000). "Mössbauer Study of the MoFe Protein of Nitrogenase from Azotobacter vinelandii Using Selective 57Fe Enrichment of the M-Centers". Journal of the American Chemical Society. 122 (20): 4926–4936. doi:10.1021/ja000254k.
^ Lukoyanov D, Barney BM, Dean DR, Seefeldt LC, Hoffman BM (January 2007). "Connecting nitrogenase intermediates with the kinetic scheme for N2 reduction by a relaxation protocol and identification of the N2 binding state". Proceedings of the National Academy of Sciences of the United States of America. 104 (5): 1451–5. doi:10.1073/pnas.0610975104. PMC 1785236. PMID 17251348.
^ ab Igarashi RY, Laryukhin M, Dos Santos PC, Lee HI, Dean DR, Seefeldt LC, Hoffman BM (May 2005). "Trapping H- bound to the nitrogenase FeMo-cofactor active site during H2 evolution: characterization by ENDOR spectroscopy". Journal of the American Chemical Society. 127 (17): 6231–41. doi:10.1021/ja043596p. PMID 15853328.
^ Doan PE, Telser J, Barney BM, Igarashi RY, Dean DR, Seefeldt LC, Hoffman BM (November 2011). "57Fe ENDOR spectroscopy and 'electron inventory' analysis of the nitrogenase E4 intermediate suggest the metal-ion core of FeMo-cofactor cycles through only one redox couple". Journal of the American Chemical Society. 133 (43): 17329–40. doi:10.1021/ja205304t. PMC 3232045. PMID 21980917.
^ abcd Neese F (December 2005). "The Yandulov/Schrock cycle and the nitrogenase reaction: pathways of nitrogen fixation studied by density functional theory". Angewandte Chemie. 45 (2): 196–9. doi:10.1002/anie.200502667. PMID 16342309.
^ abcd Hinnemann B, Nørskov JK (2008). "Catalysis by Enzymes: The Biological Ammonia Synthesis". Topics in Catalysis. 37 (1): 55–70. doi:10.1007/s11244-006-0002-0.
^ ab Schrock RR (December 2005). "Catalytic reduction of dinitrogen to ammonia at a single molybdenum center". Accounts of Chemical Research. 38 (12): 955–62. doi:10.1021/ar0501121. PMC 2551323. PMID 16359167.
^ ab Rodriguez MM, Bill E, Brennessel WW, Holland PL (November 2011). "N₂reduction and hydrogenation to ammonia by a molecular iron-potassium complex". Science. 334 (6057): 780–3. doi:10.1126/science.1211906. PMC 3218428. PMID 22076372.
^ ab Anderson JS, Rittle J, Peters JC (September 2013). "Catalytic conversion of nitrogen to ammonia by an iron model complex". Nature. 501 (7465): 84–7. doi:10.1038/nature12435. PMC 3882122. PMID 24005414.
^ Chatt J, Dilworth JR, Richards RL (1978). "Recent advances in chemistry of nitrogen-fixation". Chem. Rev. 78: 589–625. doi:10.1021/cr60316a001.
^ Murakami J, Yamaguchi W (2012-05-14). "Reduction of N2 by supported tungsten clusters gives a model of the process by nitrogenase". Scientific Reports. 2: 407. doi:10.1038/srep00407. PMC 3350986. PMID 22586517.
^ Dilworth MJ, Eady RR (July 1991). "Hydrazine is a product of dinitrogen reduction by the vanadium-nitrogenase from Azotobacter chroococcum". The Biochemical Journal. 277 (2): 465–8. doi:10.1042/bj2770465. PMC 1151257. PMID 1859374.
^ Hageman RV, Burris RH (June 1978). "Nitrogenase and nitrogenase reductase associate and dissociate with each catalytic cycle". Proceedings of the National Academy of Sciences of the United States of America. 75 (6): 2699–702. doi:10.1073/pnas.75.6.2699. PMC 392630. PMID 275837.
^ ab Georgiadis MM, Komiya H, Chakrabarti P, Woo D, Kornuc JJ, Rees DC (September 1992). "Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii". Science. 257 (5077): 1653–9. doi:10.1126/science.1529353. PMID 1529353.
^ Seefeldt LC, Morgan TV, Dean DR, Mortenson LE (April 1992). "Mapping the site(s) of MgATP and MgADP interaction with the nitrogenase of Azotobacter vinelandii. Lysine 15 of the iron protein plays a major role in MgATP interaction". The Journal of Biological Chemistry. 267 (10): 6680–8. PMID 1313018.
^ Ryle MJ, Lanzilotta WN, Mortenson LE, Watt GD, Seefeldt LC (June 1995). "Evidence for a central role of lysine 15 of Azotobacter vinelandii nitrogenase iron protein in nucleotide binding and protein conformational changes". The Journal of Biological Chemistry. 270 (22): 13112–7. doi:10.1074/jbc.270.22.13112. PMID 7768906.
^ ab Wolle D, Dean DR, Howard JB (November 1992). "Nucleotide-iron-sulfur cluster signal transduction in the nitrogenase iron-protein: the role of Asp125". Science. 258 (5084): 992–5. doi:10.1126/science.1359643. PMID 1359643.
^ "Nuclear Magnetic Resonance Spectra of Adenosine Di- and Triphosphate". Journal of Biological Chemistry. 237: 176–181. 1962.
^ ab Chen L, Gavini N, Tsuruta H, Eliezer D, Burgess BK, Doniach S, Hodgson KO (February 1994). "MgATP-induced conformational changes in the iron protein from Azotobacter vinelandii, as studied by small-angle x-ray scattering". The Journal of Biological Chemistry. 269 (5): 3290–4. PMID 8106367.
^ Seefeldt LC, Dance IG, Dean DR (February 2004). "Substrate interactions with nitrogenase: Fe versus Mo". Biochemistry. 43 (6): 1401–9. doi:10.1021/bi036038g. PMID 14769015.
^ Oelze J (October 2000). "Respiratory protection of nitrogenase in Azotobacter species: is a widely held hypothesis unequivocally supported by experimental evidence?". FEMS Microbiology Reviews. 24 (4): 321–33. doi:10.1111/j.1574-6976.2000.tb00545.x. PMID 10978541.
^ ab Rivera-Ortiz JM, Burris RH (August 1975). "Interactions among substrates and inhibitors of nitrogenase". Journal of Bacteriology. 123 (2): 537–45. PMC 235759. PMID 1150625.
^ ab Schrauzer GN (August 1975). "Nonenzymatic simulation of nitrogenase reactions and the mechanism of biological nitrogen fixation". Angewandte Chemie. 14 (8): 514–22. doi:10.1002/anie.197505141. PMID 810048.
^ abc Seefeldt LC, Rasche ME, Ensign SA (April 1995). "Carbonyl sulfide and carbon dioxide as new substrates, and carbon disulfide as a new inhibitor, of nitrogenase". Biochemistry. 34 (16): 5382–9. doi:10.1021/bi00016a009. PMID 7727396.
^ abc Rasche ME, Seefeldt LC (July 1997). "Reduction of thiocyanate, cyanate, and carbon disulfide by nitrogenase: kinetic characterization and EPR spectroscopic analysis". Biochemistry. 36 (28): 8574–85. doi:10.1021/bi970217e. PMID 9214303.
^ Guth JH, Burris RH (October 1983). "Inhibition of nitrogenase-catalyzed NH3 formation by H2". Biochemistry. 22 (22): 5111–22. doi:10.1021/bi00291a010. PMID 6360203.
^ Li J, Goldschmidt-Clermont M, Timko MP (December 1993). "Chloroplast-encoded chlB is required for light-independent protochlorophyllide reductase activity in Chlamydomonas reinhardtii". The Plant Cell. 5 (12): 1817–29. doi:10.1105/tpc.5.12.1817. PMC 160407. PMID 8305874.
^ ab Staples CR, Lahiri S, Raymond J, Von Herbulis L, Mukhophadhyay B, Blankenship RE (October 2007). "Expression and association of group IV nitrogenase NifD and NifH homologs in the non-nitrogen-fixing archaeon Methanocaldococcus jannaschii". Journal of Bacteriology. 189 (20): 7392–8. doi:10.1128/JB.00876-07. PMC 2168459. PMID 17660283.
^ Raymond J, Siefert JL, Staples CR, Blankenship RE (March 2004). "The natural history of nitrogen fixation". Molecular Biology and Evolution. 21 (3): 541–54. doi:10.1093/molbev/msh047. PMID 14694078.
^ Dilworth MJ (October 1966). "Acetylene reduction by nitrogen-fixing preparations from Clostridium pasteurianum". Biochimica et Biophysica Acta. 127 (2): 285–94. doi:10.1016/0304-4165(66)90383-7. PMID 5964974.
^ Sims GK, Dunigan EP (1984). "Diurnal and seasonal variations in nitrogenase activity (C2H2 reduction) of rice roots". Soil Biology and Biochemistry. 16: 15–18. doi:10.1016/0038-0717(84)90118-4.
Further reading[edit]
.mw-parser-output .refbeginfont-size:90%;margin-bottom:0.5em.mw-parser-output .refbegin-hanging-indents>ullist-style-type:none;margin-left:0.mw-parser-output .refbegin-hanging-indents>ul>li,.mw-parser-output .refbegin-hanging-indents>dl>ddmargin-left:0;padding-left:3.2em;text-indent:-3.2em;list-style:none.mw-parser-output .refbegin-100font-size:100%
Zumft WG, Mortenson LE (March 1975). "The nitrogen-fixing complex of bacteria". Biochimica et Biophysica Acta. 416 (1): 1–52. doi:10.1016/0304-4173(75)90012-9. PMID 164247.
External links[edit]
 Media related to Nitrogenase at Wikimedia Commons
Media related to Nitrogenase at Wikimedia Commons
Categories:
- EC 1.18.6
- Iron-sulfur proteins
- Nitrogen cycle
(window.RLQ=window.RLQ||).push(function()mw.config.set("wgPageParseReport":"limitreport":"cputime":"2.876","walltime":"4.287","ppvisitednodes":"value":3755,"limit":1000000,"ppgeneratednodes":"value":0,"limit":1500000,"postexpandincludesize":"value":153534,"limit":2097152,"templateargumentsize":"value":4093,"limit":2097152,"expansiondepth":"value":15,"limit":40,"expensivefunctioncount":"value":5,"limit":500,"unstrip-depth":"value":1,"limit":20,"unstrip-size":"value":158407,"limit":5000000,"entityaccesscount":"value":3,"limit":400,"timingprofile":["100.00% 3815.814 1 -total"," 62.31% 2377.553 1 Template:Reflist"," 54.93% 2096.107 44 Template:Cite_journal"," 6.89% 263.013 1 Template:Chem"," 6.53% 249.231 1 Template:Chem/link"," 6.46% 246.576 1 Template:Enzyme"," 6.38% 243.554 2 Template:Chem/atom"," 6.37% 243.250 2 Template:Infobox"," 6.37% 243.231 4 Template:Citation_needed"," 5.58% 212.802 4 Template:Fix"],"scribunto":"limitreport-timeusage":"value":"1.909","limit":"10.000","limitreport-memusage":"value":5780858,"limit":52428800,"cachereport":"origin":"mw1244","timestamp":"20190310181537","ttl":2592000,"transientcontent":false););"@context":"https://schema.org","@type":"Article","name":"Nitrogenase","url":"https://en.wikipedia.org/wiki/Nitrogenase","sameAs":"http://www.wikidata.org/entity/Q410381","mainEntity":"http://www.wikidata.org/entity/Q410381","author":"@type":"Organization","name":"Contributors to Wikimedia projects","publisher":"@type":"Organization","name":"Wikimedia Foundation, Inc.","logo":"@type":"ImageObject","url":"https://www.wikimedia.org/static/images/wmf-hor-googpub.png","datePublished":"2004-10-18T18:38:58Z","dateModified":"2019-02-10T00:19:07Z","image":"https://upload.wikimedia.org/wikipedia/commons/9/9b/Nitrogenase.png","headline":"enzyme"(window.RLQ=window.RLQ||).push(function()mw.config.set("wgBackendResponseTime":119,"wgHostname":"mw1328"););

